T Cell biology laboratory
Our long-term research interest is to characterize molecular mechanisms that regulate the development and function of T cells. In previous studies we characterized in detail the transcriptional control of the Cd8 gene expression and demonstrated that an active state of Cd8a and Cd8b gene expression is epigenetically maintained during the development of DP thymocytes. We further identified that the transcription factor MAZR/PATZ1 is an important regulator of CD8 expression, CD4/CD8 lineage development and the development of FOXP3+ regulatory T cells. In preclinical mouse models we also identified that histone deacetylase 1 (HDAC1) in T cells is a key factor in driving T cell-mediated autoimmune diseases such as experimental autoimmune encephalomyelitis, collagen-induced arthritis and adoptive transfer colitis. We also showed that HDAC1/HDAC2 control the lineage integrity of CD4+ T cells and restrain the induction of CD4+ T cells with cytotoxic activity (CD4 CTL). In ongoing studies we aim to understand transcriptional networks that control peripheral T cell differentiation. We are dissecting MAZR/PATZ1 function and analyzing the role of the transcriptional adapter protein NCOR1. Moreover, we investigate how histone deacetylases regulate T cell function beyond chromatin-mediated control of gene expression and study whether selective HDAC inhibition and/or targeting HDAC-specific pathways is a promising treatment strategy for T-cell-mediated diseases.
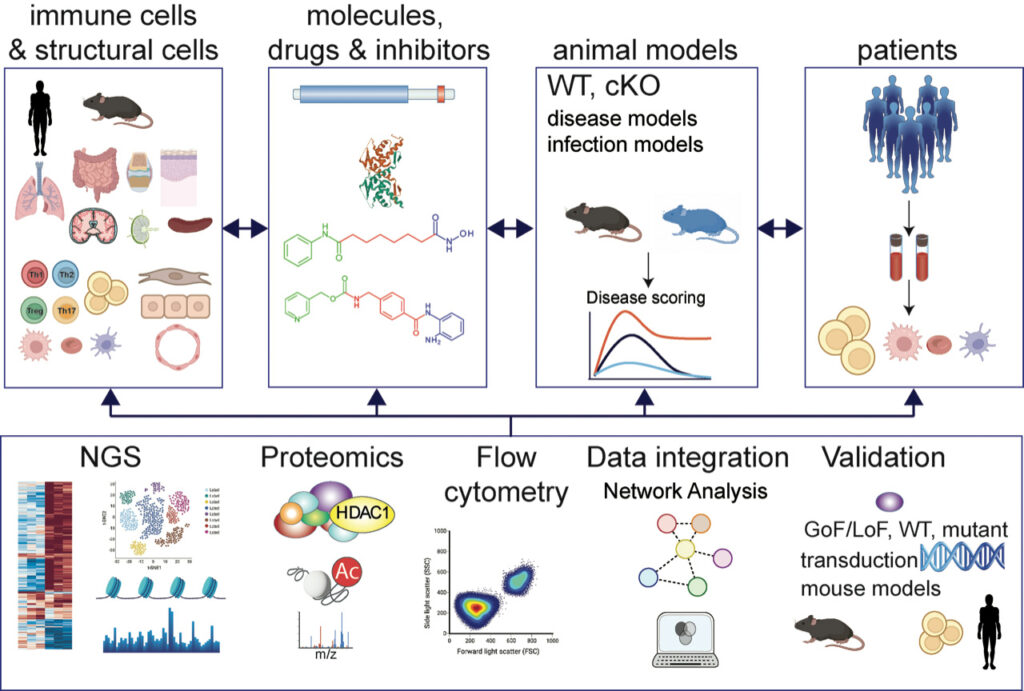
We are embedded in a strong collaborative research environment. The experimental strategies to address our research interests include multi-color flow-cytometry, a variety of immunological tools, biochemical and molecular approaches, retroviral-mediated gene transduction into T cells and hematopoietic stem cells, next generation sequencing and mouse molecular genetics tools combined with preclinical models for autoimmune diseases and infections.
Research topics
The role of histone deacetylases in the control of T cell-mediated immunity
The differentiation of T cell subsets and their acquisition of effector functions are accompanied by changes in gene expression programs, which in part are regulated and maintained by epigenetic processes. Histone deacetylases (HDACs) and histone acetyltransferases (HATs) are key epigenetic regulators that function by mediating dynamic changes in the acetylation of histones at lysine residues and thus regulating gene expression. In addition, in recent years many non-histone targets were identified and reversible acetylation affects their functional properties (such as protein-protein and protein-DNA interactions, protein stability, enzymatic activity, intracellular localization) and is also linked to metabolism. This demonstrates that HAT/HDACs function beyond chromatin-mediated control of gene expression and that lysine acetylation is an important post-translational modification likely to be comparable with protein phosphorylation. Eighteen members of the HDAC family have been identified in mammalian organisms, however dissecting individual roles for each HDACs in specific cell lineages and tissues remains a major scientific challenge. We are analyzing the role of HDAC1, HDAC2 and other members of the HDAC family in T cells and T cell-mediated immune diseases. These studies are performed in the framework of a special research program (SFB) “HDACs as regulators of T cell-mediated immunity in health and disease”, which is a newly established research network funded by the Austrian Science Fund (www.meduniwien.ac.at/HIT). The SFB is formed by an interdisciplinary consortium of nine research groups, of which eight are located in Vienna and one in Salzburg. Our publications related to HDAC function in T cells can be found <here>.
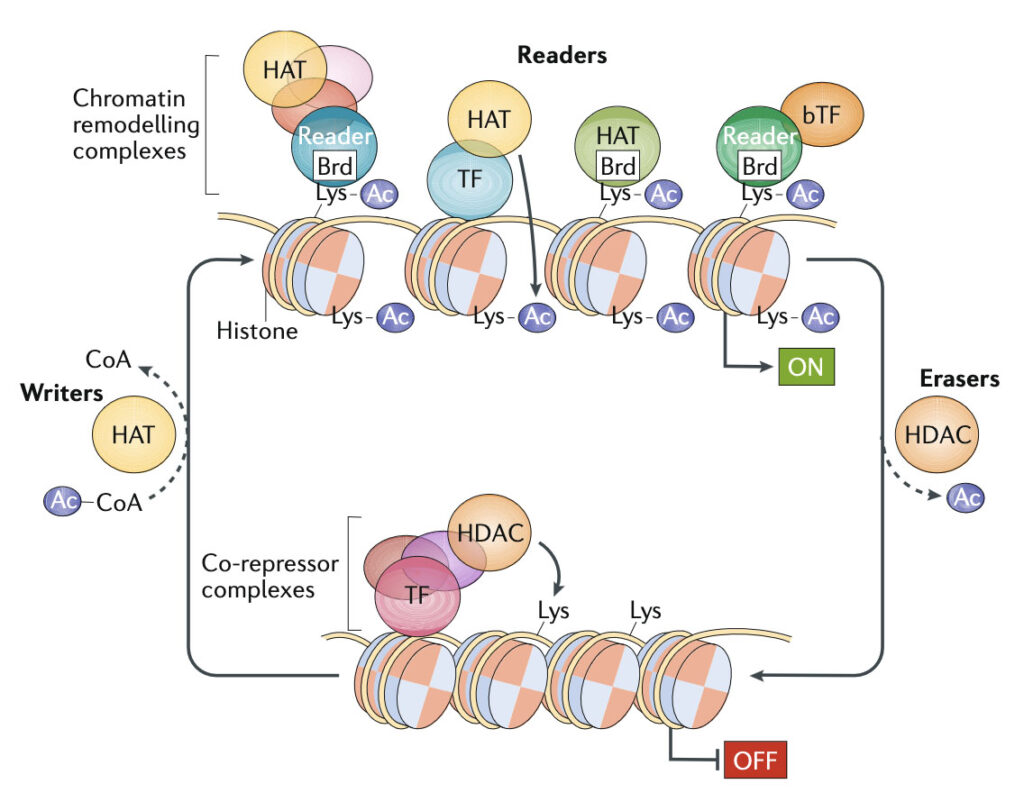
In general, the active state of genes correlates with increased presence of acetylated histones (Ac). Reversible lysine acetylation of histones mediated by histone acetyltransferases (HATs) has two major consequences. First, it reduces electrostatic interactions between DNA and histone proteins, resulting in a more open chromatin structure and thus allowing the recruitment of gene-specific transcription factors (TFs). Second, it attracts specific epigenetic reader proteins that contain bromodomains (Brds). These readers might recruit the basal TFs (bTFs) or chromatin remodelling complexes. Transcriptional repressors recruit co-repressor complexes that contain histone deacetylases (HDACs), leading to the deacetylation of histones and thus to a closed chromatin configuration. For simplicity, other histone marks such as methylation or phosphorylation have been omitted. Figure taken from Ellmeier and Seiser, 2018, Nature Reviews Immunology.
Molecular analysis of the transcription factor MAZR/PATZ1
The BTB domain-containing zinc finger protein MAZR/PATZ1 is a key regulator of many biological processes. Loss of MAZR/PATZ1 leads to embryonic lethality and altered expression of MAZR/PATZ1 is linked with tumorigenesis. We have identified that MAZR/PATZ1 is essential for the transcriptional regulator of CD8 expression (Bilic et al., 2006, Nat Immunol) and further demonstrated that MAZR/PATZ1 is part of the transcription factor network regulating CD4/CD8 cell fate choice of DP thymocytes by repressing ThPOK expression in MHC class I-signaled DP thymocytes (Sakaguchi et al., 2010, Nat Immunol; Sakaguchi et al., J Immunol, 2015). Using mice with a conditional deletion of MAZR in the T cell lineage we uncovered a crucial role for MAZR/PATZ1 in regulating the development and function of FOXP3+ regulatory T cells (Andersen et al., 2019) and iNKT cell subset differentiation (in collaboration with the Sakaguchi group; Orola et al. 2019). In ongoing studies we further characterize MAZR/PATZ1 function in T cells and beyond. Our publications related to MAZR/PATZ1 function can be found <here>.
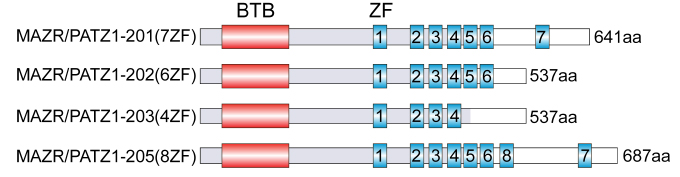
Schematic map of MAZR/PATZ1. Several isoforms that differ in the number of C-terminal Zn-fingers (ZF) are known. The BTB domain, also known as POZ domain is a protein–protein interaction motif that is present in about 200 genes in mammalian genomes and there are around 44 different ZF transcription factors that contain a BTB domain. The MAZR/PATZ1 isoforms arise due to alternative splicing. Whether the isoforms differ in their function is not known.
NCOR1 – a key regulator of Th cell differentiation
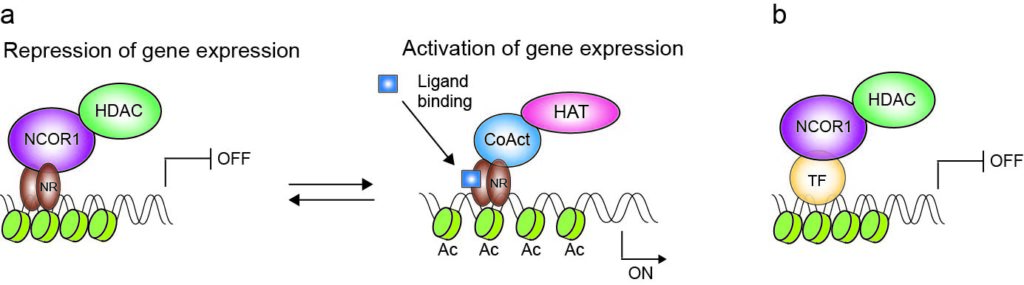
NCOR1 belongs to the important group of transcriptional cofactors that connects repressive chromatin-modifying enzymes with gene specific transcription factors. We previously demonstrated essential roles for NCOR1 during T cell development and in orchestrating transcriptional landscapes and effector function in peripheral CD4+ T cells. In ongoing studies we are addressing the role of NCOR1 in regulating T cell-mediated immunity in tissues. Our publications about NCOR1 can be found <here>.
(a) NCOR1 mediates transcriptional repression by bridging histone deacetylases (HDACs) with nuclear receptors (NR) in the absence of their ligands. Direct activation of NRs by ligand binding induces a conformational change followed by the exchange of NCOR1 corepressor complexes with coactivator complexes (CoAct). Coactivator complexes are usually associated with histone acetyltransferases (HATs), which promote transcription. (b) NCOR1 interacts also with othertranscription factors (e.g. transcriptional repressors) and thus regulates their repressive activity. Figure taken from Müller et al., 2018, J Leukoc Bio.
During the last years, our research has been supported by:
• Austrian Science Fund (FWF), stand alone projects P23641, P26193, P29790, P34407, P35372
• FWF/MedUni Wien Doktoratskolleg „Inflammation and Immunity“ (W1212)
• Horizon 2020, Marie Sklodowska Curie Innovative Training Network „ENLIGHT-TEN“
• Vienna Science and Technology Fund (WWTF), Life Sciences Call 2009: Project: Epigenetic Regulation of T Cell Development and Function (LS09-031), awared to Wilfried Ellmeier and Christian Seiser
• FWF/MedUni Wien Doc.Fund PhD program „TissueHome“ (DOC32)
• Austrian Science Fund (FWF), Special Research Program (SFB) F-70 („HDACs as regulators of T cell-mediated immunity„)
• Horizon 2020, Marie Sklodowska Curie Innovative Training Network ENLIGHT-TEN+
• FWF/MedUni Wien Doc.Fund PhD program „SHIELD“ (DOC188)
